Medical image datasets#
TorchIO offers tools to easily download publicly available datasets from different institutions and modalities.
The interface is similar to torchvision.datasets.
If you use any of them, please visit the corresponding website (linked in each description) and make sure you comply with any data usage agreement and you acknowledge the corresponding authors’ publications.
If you would like to add a dataset here, please open a discussion on the GitHub repository:
DiscussIXI#
The Information eXtraction from Images (IXI) dataset contains “nearly 600 MR images from normal, healthy subjects”, including “T1, T2 and PD-weighted images, MRA images and Diffusion-weighted images (15 directions)”.
Note
This data is made available under the Creative Commons CC BY-SA 3.0 license. If you use it please acknowledge the source of the IXI data, e.g. the IXI website.
IXI#
- class torchio.datasets.ixi.IXI(root: str | Path, transform: Transform | None = None, download: bool = False, modalities: Sequence[str] = ('T1', 'T2'), **kwargs)[source]#
Bases:
SubjectsDatasetFull IXI dataset.
- Parameters:
root – Root directory to which the dataset will be downloaded.
transform – An instance of
Transform.download – If set to
True, will download the data intoroot.modalities – List of modalities to be downloaded. They must be in
('T1', 'T2', 'PD', 'MRA', 'DTI').
Warning
The size of this dataset is multiple GB. If you set
downloadtoTrue, it will take some time to be downloaded if it is not already present.Example
>>> import torchio as tio >>> transforms = [ ... tio.ToCanonical(), # to RAS ... tio.Resample((1, 1, 1)), # to 1 mm iso ... ] >>> ixi_dataset = tio.datasets.IXI( ... 'path/to/ixi_root/', ... modalities=('T1', 'T2'), ... transform=tio.Compose(transforms), ... download=True, ... ) >>> print('Number of subjects in dataset:', len(ixi_dataset)) # 577 >>> sample_subject = ixi_dataset[0] >>> print('Keys in subject:', tuple(sample_subject.keys())) # ('T1', 'T2') >>> print('Shape of T1 data:', sample_subject['T1'].shape) # [1, 180, 268, 268] >>> print('Shape of T2 data:', sample_subject['T2'].shape) # [1, 241, 257, 188]
IXITiny#
- class torchio.datasets.ixi.IXITiny(root: str | Path, transform: Transform | None = None, download: bool = False, **kwargs)[source]#
Bases:
SubjectsDatasetThis is the dataset used in the main notebook. It is a tiny version of IXI, containing 566 \(T_1\)-weighted brain MR images and their corresponding brain segmentations, all with size \(83 \times 44 \times 55\).
It can be used as a medical image MNIST.
- Parameters:
root – Root directory to which the dataset will be downloaded.
transform – An instance of
Transform.download – If set to
True, will download the data intoroot.
EPISURG#
EPISURG#
- class torchio.datasets.episurg.EPISURG(root: str | Path, transform: Transform | None = None, download: bool = False, **kwargs)[source]#
Bases:
SubjectsDatasetEPISURG is a clinical dataset of \(T_1\)-weighted MRI from 430 epileptic patients who underwent resective brain surgery at the National Hospital of Neurology and Neurosurgery (Queen Square, London, United Kingdom) between 1990 and 2018.
The dataset comprises 430 postoperative MRI. The corresponding preoperative MRI is present for 268 subjects.
Three human raters segmented the resection cavity on partially overlapping subsets of EPISURG.
If you use this dataset for your research, you agree with the Data use agreement presented at the EPISURG entry on the UCL Research Data Repository and you must cite the corresponding publications.
- Parameters:
root – Root directory to which the dataset will be downloaded.
transform – An instance of
Transform.download – If set to
True, will download the data intoroot.
Warning
The size of this dataset is multiple GB. If you set
downloadtoTrue, it will take some time to be downloaded if it is not already present.- get_labeled() SubjectsDataset[source]#
Get dataset from subjects with manual annotations.
- get_paired() SubjectsDataset[source]#
Get dataset from subjects with pre- and post-op MRI.
- get_unlabeled() SubjectsDataset[source]#
Get dataset from subjects without manual annotations.
Kaggle datasets#
RSNAMICCAI#
- class torchio.datasets.rsna_miccai.RSNAMICCAI(root_dir: str | Path, train: bool = True, ignore_empty: bool = True, modalities: Sequence[str] = ('T1w', 'T1wCE', 'T2w', 'FLAIR'), **kwargs)[source]#
Bases:
SubjectsDatasetRSNA-MICCAI Brain Tumor Radiogenomic Classification challenge dataset.
This is a helper class for the dataset used in the RSNA-MICCAI Brain Tumor Radiogenomic Classification challenge hosted on kaggle. The dataset must be downloaded before instantiating this class (as opposed to, e.g.,
torchio.datasets.IXI).This kaggle kernel includes a usage example including preprocessing of all the scans.
If you reference or use the dataset in any form, include the following citation:
U.Baid, et al., “The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification”, arXiv:2107.02314, 2021.
- Parameters:
root_dir – Directory containing the dataset (
traindirectory,testdirectory, etc.).train – If
True, thetrainset will be used. Otherwise thetestset will be used.ignore_empty – If
True, the three subjects flagged as “presenting issues” (empty images) by the challenge organizers will be ignored. The subject IDs are00109,00123and00709.
Example
>>> import torchio as tio >>> from subprocess import call >>> call('kaggle competitions download -c rsna-miccai-brain-tumor-radiogenomic-classification'.split()) >>> root_dir = 'rsna-miccai-brain-tumor-radiogenomic-classification' >>> train_set = tio.datasets.RSNAMICCAI(root_dir, train=True) >>> test_set = tio.datasets.RSNAMICCAI(root_dir, train=False) >>> len(train_set), len(test_set) (582, 87)
RSNACervicalSpineFracture#
- class torchio.datasets.rsna_spine_fracture.RSNACervicalSpineFracture(root_dir: str | Path, add_segmentations: bool = False, add_bounding_boxes: bool = False, **kwargs)[source]#
Bases:
SubjectsDatasetRSNA 2022 Cervical Spine Fracture Detection dataset.
This is a helper class for the dataset used in the RSNA 2022 Cervical Spine Fracture Detection hosted on kaggle. The dataset must be downloaded before instantiating this class.
MNI#
ICBM2009CNonlinearSymmetric#
- class torchio.datasets.mni.ICBM2009CNonlinearSymmetric(load_4d_tissues: bool = True)[source]#
Bases:
SubjectMNIICBM template.
More information can be found in the website.
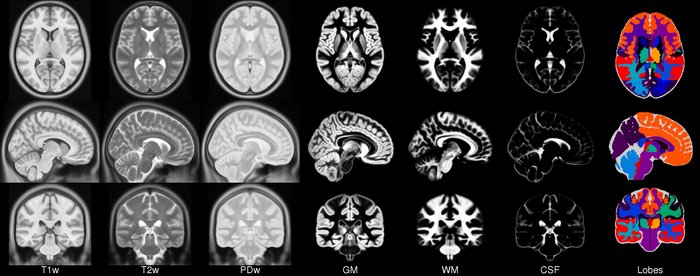
- Parameters:
load_4d_tissues – If
True, the tissue probability maps will be loaded together into a 4D image. Otherwise, they will be loaded into independent images.
Example
>>> import torchio as tio >>> icbm = tio.datasets.ICBM2009CNonlinearSymmetric() >>> icbm ICBM2009CNonlinearSymmetric(Keys: ('t1', 'eyes', 'face', 'brain', 't2', 'pd', 'tissues'); images: 7) >>> icbm = tio.datasets.ICBM2009CNonlinearSymmetric(load_4d_tissues=False) >>> icbm ICBM2009CNonlinearSymmetric(Keys: ('t1', 'eyes', 'face', 'brain', 't2', 'pd', 'gm', 'wm', 'csf'); images: 9)
Colin27#
(Source code, png)

Pediatric#
- class torchio.datasets.mni.Pediatric(years, symmetric=False)[source]#
Bases:
SubjectMNIMNI pediatric atlases.
See the MNI website for more information.
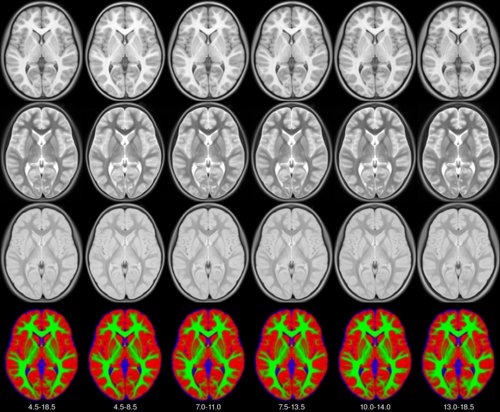
- Parameters:
years – Tuple of 2 ages. Possible values are:
(4.5, 18.5),(4.5, 8.5),(7, 11),(7.5, 13.5),(10, 14)and(13, 18.5).symmetric – If
True, the left-right symmetric templates will be used. Else, the asymmetric (natural) templates will be used.
(Source code, png)

Sheep#
(Source code, png)

BITE3#
- class torchio.datasets.bite.BITE3(root: str | Path, transform: Transform | None = None, download: bool = False, **kwargs)[source]#
Bases:
BITEPre- and post-resection MR images in BITE.
The goal of BITE is to share in vivo medical images of patients wtith brain tumors to facilitate the development and validation of new image processing algorithms.
Please check the BITE website for more information and acknowledgments instructions.
- Parameters:
root – Root directory to which the dataset will be downloaded.
transform – An instance of
Transform.download – If set to
True, will download the data intoroot.
ITK-SNAP#
BrainTumor#
(Source code, png)

T1T2#
(Source code, png)

AorticValve#
(Source code, png)

3D Slicer#
Slicer#
- class torchio.datasets.slicer.Slicer(name='MRHead')[source]#
Bases:
_RawSubjectCopySubjectSample data provided by 3D Slicer.
See the Slicer wiki for more information.
For information about licensing and permissions, check the Sample Data module.
- Parameters:
name – One of the keys in
torchio.datasets.slicer.URLS_DICT.
(Source code, png)

FPG#
- class torchio.datasets.fpg.FPG(load_all: bool = False)[source]#
Bases:
_RawSubjectCopySubject3T \(T_1\)-weighted brain MRI and corresponding parcellation.
- Parameters:
load_all – If
True, three more images will be loaded: a \(T_2\)-weighted MRI, a diffusion MRI and a functional MRI.
(Source code, png)

(Source code, png)

MedMNIST#
- class torchio.datasets.medmnist.OrganMNIST3D(split, **kwargs)[source]#
Bases:
MedMNIST3D MedMNIST v2 datasets.
Datasets from MedMNIST v2: A Large-Scale Lightweight Benchmark for 2D and 3D Biomedical Image Classification.
Please check the MedMNIST website for more information, inclusing the license.
- Parameters:
split – Dataset split. Should be
'train','val'or'test'.
(Source code, png)

- class torchio.datasets.medmnist.NoduleMNIST3D(split, **kwargs)[source]#
Bases:
MedMNIST3D MedMNIST v2 datasets.
Datasets from MedMNIST v2: A Large-Scale Lightweight Benchmark for 2D and 3D Biomedical Image Classification.
Please check the MedMNIST website for more information, inclusing the license.
- Parameters:
split – Dataset split. Should be
'train','val'or'test'.
(Source code, png)

- class torchio.datasets.medmnist.AdrenalMNIST3D(split, **kwargs)[source]#
Bases:
MedMNIST3D MedMNIST v2 datasets.
Datasets from MedMNIST v2: A Large-Scale Lightweight Benchmark for 2D and 3D Biomedical Image Classification.
Please check the MedMNIST website for more information, inclusing the license.
- Parameters:
split – Dataset split. Should be
'train','val'or'test'.
(Source code, png)

- class torchio.datasets.medmnist.FractureMNIST3D(split, **kwargs)[source]#
Bases:
MedMNIST3D MedMNIST v2 datasets.
Datasets from MedMNIST v2: A Large-Scale Lightweight Benchmark for 2D and 3D Biomedical Image Classification.
Please check the MedMNIST website for more information, inclusing the license.
- Parameters:
split – Dataset split. Should be
'train','val'or'test'.
(Source code, png)

- class torchio.datasets.medmnist.VesselMNIST3D(split, **kwargs)[source]#
Bases:
MedMNIST3D MedMNIST v2 datasets.
Datasets from MedMNIST v2: A Large-Scale Lightweight Benchmark for 2D and 3D Biomedical Image Classification.
Please check the MedMNIST website for more information, inclusing the license.
- Parameters:
split – Dataset split. Should be
'train','val'or'test'.
(Source code, png)

- class torchio.datasets.medmnist.SynapseMNIST3D(split, **kwargs)[source]#
Bases:
MedMNIST3D MedMNIST v2 datasets.
Datasets from MedMNIST v2: A Large-Scale Lightweight Benchmark for 2D and 3D Biomedical Image Classification.
Please check the MedMNIST website for more information, inclusing the license.
- Parameters:
split – Dataset split. Should be
'train','val'or'test'.
(Source code, png)
