seaborn.ecdfplot#
- seaborn.ecdfplot(data=None, *, x=None, y=None, hue=None, weights=None, stat='proportion', complementary=False, palette=None, hue_order=None, hue_norm=None, log_scale=None, legend=True, ax=None, **kwargs)#
Plot empirical cumulative distribution functions.
An ECDF represents the proportion or count of observations falling below each unique value in a dataset. Compared to a histogram or density plot, it has the advantage that each observation is visualized directly, meaning that there are no binning or smoothing parameters that need to be adjusted. It also aids direct comparisons between multiple distributions. A downside is that the relationship between the appearance of the plot and the basic properties of the distribution (such as its central tendency, variance, and the presence of any bimodality) may not be as intuitive.
More information is provided in the user guide.
- Parameters:
- data
pandas.DataFrame,numpy.ndarray, mapping, or sequence Input data structure. Either a long-form collection of vectors that can be assigned to named variables or a wide-form dataset that will be internally reshaped.
- x, yvectors or keys in
data Variables that specify positions on the x and y axes.
- huevector or key in
data Semantic variable that is mapped to determine the color of plot elements.
- weightsvector or key in
data If provided, weight the contribution of the corresponding data points towards the cumulative distribution using these values.
- stat{{“proportion”, “percent”, “count”}}
Distribution statistic to compute.
- complementarybool
If True, use the complementary CDF (1 - CDF)
- palettestring, list, dict, or
matplotlib.colors.Colormap Method for choosing the colors to use when mapping the
huesemantic. String values are passed tocolor_palette(). List or dict values imply categorical mapping, while a colormap object implies numeric mapping.- hue_ordervector of strings
Specify the order of processing and plotting for categorical levels of the
huesemantic.- hue_normtuple or
matplotlib.colors.Normalize Either a pair of values that set the normalization range in data units or an object that will map from data units into a [0, 1] interval. Usage implies numeric mapping.
- log_scalebool or number, or pair of bools or numbers
Set axis scale(s) to log. A single value sets the data axis for any numeric axes in the plot. A pair of values sets each axis independently. Numeric values are interpreted as the desired base (default 10). When
NoneorFalse, seaborn defers to the existing Axes scale.- legendbool
If False, suppress the legend for semantic variables.
- ax
matplotlib.axes.Axes Pre-existing axes for the plot. Otherwise, call
matplotlib.pyplot.gca()internally.- kwargs
Other keyword arguments are passed to
matplotlib.axes.Axes.plot().
- data
- Returns:
matplotlib.axes.AxesThe matplotlib axes containing the plot.
See also
displotFigure-level interface to distribution plot functions.
histplotPlot a histogram of binned counts with optional normalization or smoothing.
kdeplotPlot univariate or bivariate distributions using kernel density estimation.
rugplotPlot a tick at each observation value along the x and/or y axes.
Examples
Plot a univariate distribution along the x axis:
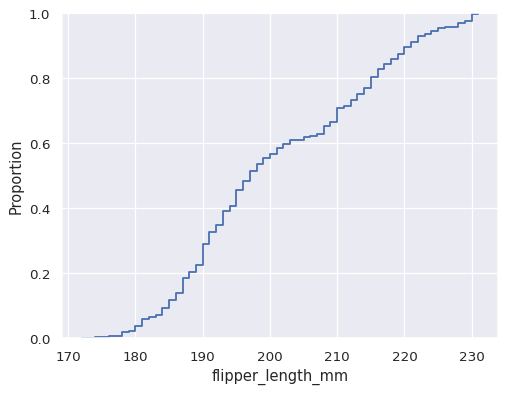
penguins = sns.load_dataset("penguins") sns.ecdfplot(data=penguins, x="flipper_length_mm")
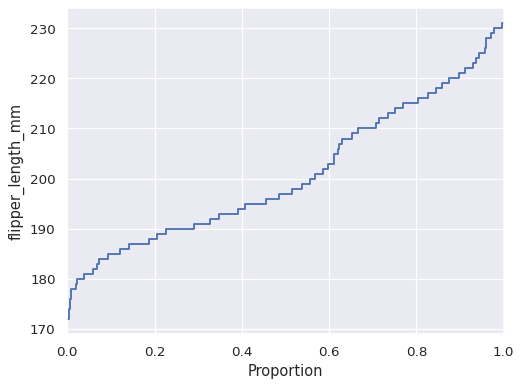
Flip the plot by assigning the data variable to the y axis:
sns.ecdfplot(data=penguins, y="flipper_length_mm")
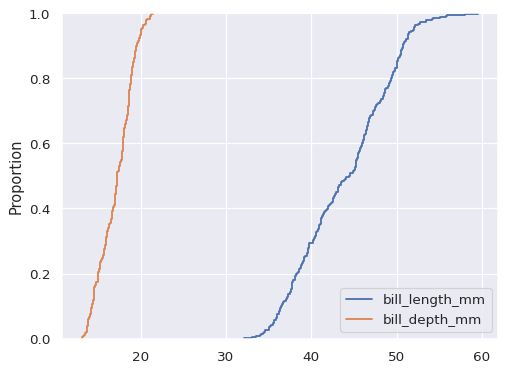
If neither
xnoryis assigned, the dataset is treated as wide-form, and a histogram is drawn for each numeric column:sns.ecdfplot(data=penguins.filter(like="bill_", axis="columns"))
You can also draw multiple histograms from a long-form dataset with hue mapping:
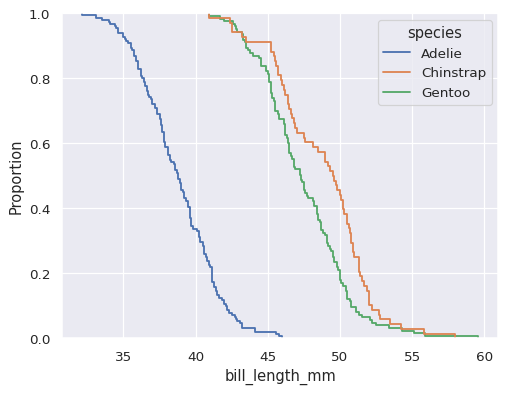
sns.ecdfplot(data=penguins, x="bill_length_mm", hue="species")
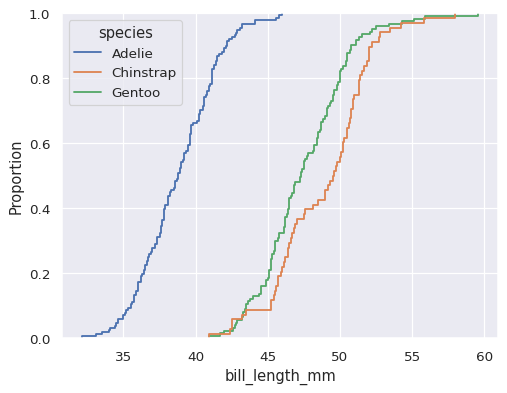
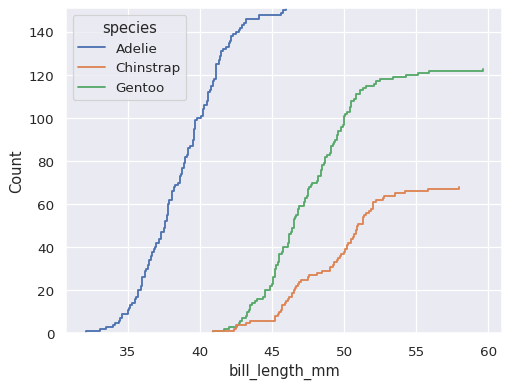
The default distribution statistic is normalized to show a proportion, but you can show absolute counts or percents instead:
sns.ecdfplot(data=penguins, x="bill_length_mm", hue="species", stat="count")
It’s also possible to plot the empirical complementary CDF (1 - CDF):
sns.ecdfplot(data=penguins, x="bill_length_mm", hue="species", complementary=True)